Profiling rare & aggressive subpopulations of cancer cells
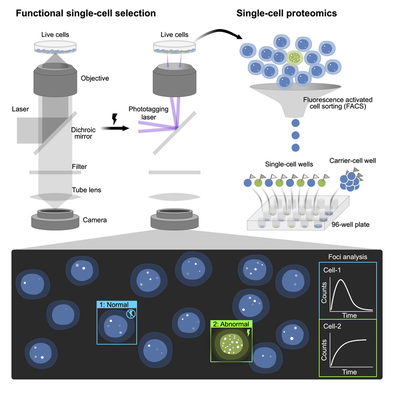
Isolation of target cells from a heterogeneous pool is technically difficult when the selection criterion is complex, such as dynamic response over time or morphological features. The majority of technologies for single-cell separation highly rely on static fluorescence or snapshot fluorescent signals, which limits the chance to identify functional and rare single cells. In addition, cancer cell subtyping is currently difficult because of a lack of reliable biomarkers for cancer cell subtypes. Metastatic cells, as an example, don’t have biomarkers that are robust across different patients and tumor types. We addressed this problem by combining our UFO microscope, a photoselection technology and an automatic pattern-recognition algorithm to identify and isolate desired cells from cultures based on complex cellular features or dynamics.
The whole pipeline is called functional single cell selection and isolation (fSCS). fSCS-isolated cells can then be subjected to single cell sequencing (FUNseq) or single cell proteomic profiling (FUNpro).
* Investigation of underlying mechanisms of cancer cells displaying aggressive migration and mesenchymal-like morphology using FUNseq
Isolation of target cells from a heterogeneous pool is technically difficult when the selection criterion is complex, such as dynamic response over time or morphological features. The majority of technologies for single-cell separation highly rely on static fluorescence or snapshot fluorescent signals, which limits the chance to identify functional and rare single cells. In addition, cancer cell subtyping is currently difficult because of a lack of reliable biomarkers for cancer cell subtypes. Metastatic cells, as an example, don’t have biomarkers that are robust across different patients and tumor types. We addressed this problem by combining our UFO microscope, a photoselection technology and an automatic pattern-recognition algorithm to identify and isolate desired cells from cultures based on complex cellular features or dynamics.
The whole pipeline is called functional single cell selection and isolation (fSCS). fSCS-isolated cells can then be subjected to single cell sequencing (FUNseq) or single cell proteomic profiling (FUNpro).
* Investigation of underlying mechanisms of cancer cells displaying aggressive migration and mesenchymal-like morphology using FUNseq
Related references:
You L.*, Su P.R.*, Betjes M*. Ghadiri Rad, R., Chou, T.C., Beerens, C., van Oosten, E., Leufkens, F., Gasecka, P., Muraro, M., van Tol, R., van Steenderen, D., Farooq, S., Hardillo, J., Baatenburg de Jong, R., Brinks, D., Chien, M.P. “Linking the genotypes and phenotypes of cancer cells in heterogenous populations via real-time optical tagging and image analysis”, Nature Biomedical Engineering, 2022 (https://doi.org/10.1038/s41551-022-00853-x)
Brinks, D., Chien, M.P. "Functional single-cell sequencing links dynamic phenotypes to their genotypes". Nature Biomedical Engineering, 2022. (https://rdcu.be/cJr8o) (https://doi.org/10.1038/s41551-022-00877-3)
Chou, T.C.*, You, L.*, Beerens, C., Feller, K.F., Storteboom, J., Chien, M.P. "Fast and Accurate Cell Tracking: a real-time cell segmentation and tracking algorithm to instantly export quantifiable cellular characteristics from big image data". Cell Reports Methods (accepted); (BioRxiv, doi: https://doi.org/10.1101/2023.01.09.523224).
You L.*, Su P.R.*, Betjes M*. Ghadiri Rad, R., Chou, T.C., Beerens, C., van Oosten, E., Leufkens, F., Gasecka, P., Muraro, M., van Tol, R., van Steenderen, D., Farooq, S., Hardillo, J., Baatenburg de Jong, R., Brinks, D., Chien, M.P. “Linking the genotypes and phenotypes of cancer cells in heterogenous populations via real-time optical tagging and image analysis”, Nature Biomedical Engineering, 2022 (https://doi.org/10.1038/s41551-022-00853-x)
Brinks, D., Chien, M.P. "Functional single-cell sequencing links dynamic phenotypes to their genotypes". Nature Biomedical Engineering, 2022. (https://rdcu.be/cJr8o) (https://doi.org/10.1038/s41551-022-00877-3)
Chou, T.C.*, You, L.*, Beerens, C., Feller, K.F., Storteboom, J., Chien, M.P. "Fast and Accurate Cell Tracking: a real-time cell segmentation and tracking algorithm to instantly export quantifiable cellular characteristics from big image data". Cell Reports Methods (accepted); (BioRxiv, doi: https://doi.org/10.1101/2023.01.09.523224).
DNA damage response and cancer biology
Understanding the complexity of cancer highly depends on an elucidation of the changes in the underlying regulatory signaling networks. As an example, resistant cancer cells have a superior ability to repair DNA damage upon chemo- or radio-therapy. This ability involves DNA-damage response (DDR) signaling pathways. We are interested in unraveling the underlying mechanisms of different DNA damage responses and their correlation with cancer cell fate.
* Dissecting the molecular mechanisms of cancer cells displaying different DNA damage repair dynamics using FUNpro
Understanding the complexity of cancer highly depends on an elucidation of the changes in the underlying regulatory signaling networks. As an example, resistant cancer cells have a superior ability to repair DNA damage upon chemo- or radio-therapy. This ability involves DNA-damage response (DDR) signaling pathways. We are interested in unraveling the underlying mechanisms of different DNA damage responses and their correlation with cancer cell fate.
* Dissecting the molecular mechanisms of cancer cells displaying different DNA damage repair dynamics using FUNpro
Related reference:
Su, P.R., You, L., Beerens, C., Bezstarosti, K., Demmers, J., Pabst, M., Kanaar, R., Hsu, C.C., Chien, M.P. “Microscopy-based single-cell proteomic profiling reveals heterogeneity in DNA damage response dynamics”, Cell Reports Methods, 2022, https://doi.org/10.1016/j.crmeth.2022.100237.
Su, P.R., You, L., Beerens, C., Bezstarosti, K., Demmers, J., Pabst, M., Kanaar, R., Hsu, C.C., Chien, M.P. “Microscopy-based single-cell proteomic profiling reveals heterogeneity in DNA damage response dynamics”, Cell Reports Methods, 2022, https://doi.org/10.1016/j.crmeth.2022.100237.
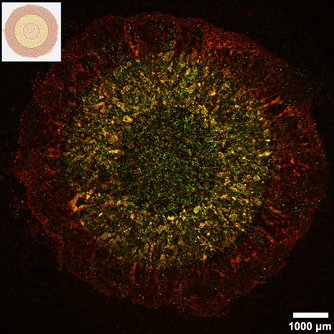
Single cell spatial -omics profiling of tumor heterogeneity
Spatial -omics profiling methods can reveal how cells interact and communicate across the cell or tissue landscape. Most currently available spatial -omics profiling techniques either have single cell resolution or in-depth profiling. We are developing a single cell spatial -omics profiling technique that can do both by combining our FUNseq technology with multiplexing methods.
* Unraveling intratumoral heterogeneity by spatially resolved profiling of cancer cells
* Unraveling intratumoral heterogeneity by spatially resolved profiling of cancer cells
Related references:
Smit, M., Feller, K.J., You, L., Chien, M.P. "Protocol for profiling intratumor heterogeneity using spatially annotated single cell sequencing." STAR Protocols. 2023, July. (DOI: 10.1016/j.xpro.2023.102447)
Smit M., Feller K., You L., Storeteboom J., Begce Y., Beerens C. Chien M.P. "Spatially annotated single cell sequencing for unraveling intratumor heterogeneity”, Frontiers in Bioengineering and Biotechnology, 2022 (https://doi.org/10.3389/fbioe.2022.829509).
Chen, T.Y., You, L. Hardillo, J.A.U., Chien, M.P. "Spatial Transcriptomic Technologies." Cells. 2023, 12(16), 2042, DOI: https://doi.org/10.3390/cells12162042.
Smit, M., Feller, K.J., You, L., Chien, M.P. "Protocol for profiling intratumor heterogeneity using spatially annotated single cell sequencing." STAR Protocols. 2023, July. (DOI: 10.1016/j.xpro.2023.102447)
Smit M., Feller K., You L., Storeteboom J., Begce Y., Beerens C. Chien M.P. "Spatially annotated single cell sequencing for unraveling intratumor heterogeneity”, Frontiers in Bioengineering and Biotechnology, 2022 (https://doi.org/10.3389/fbioe.2022.829509).
Chen, T.Y., You, L. Hardillo, J.A.U., Chien, M.P. "Spatial Transcriptomic Technologies." Cells. 2023, 12(16), 2042, DOI: https://doi.org/10.3390/cells12162042.
Machine learning-assisted cell fate prediction and target discovery
We are not only interested in investigating rare & aggressive subpopulations of cancer cells displaying tumorigenic phenotypes (like abnormal division): we also study the cells that are destined to become those aggressive or abnormal cancer cells. We employ deep learning-assisted methods to predict cell fate of individual cancer cells and single cell profile them before they develop aggressive phenotypes. In addition, we also apply (machine learning-assisted) bioinformatic analysis to identify actionable targets (biomarkers or druggable targets) that are associated with tumor metastasis, evolution or therapy resistance.